The virology pages are organized like this:
- What is a virus?
- Review of genomes, RNA, DNA and replication
- Viral genomes
- Structures of viruses
- Attachment & entry
- Viral replication
Seven types of viral genomes
Professor David Baltimore, who won the Nobel prize in medicine in 1975 for his work on retroviruses, is credited with this classification of viruses. It's often referred to as the Baltimore scheme. Baltimore didn't actually find the seventh category; that was added later, but that's how science works.
There are only* seven known types of genomes in all of virology. They differ in whether the genetic material is DNA or RNA, whether it is single- or double-stranded, and a couple of other features. We'll go through all of those below, but there is one very important principle to remember about all viruses:
The viral genome must ultimately be converted to mRNA in order to be transcribed by the ribosomes of the host cell.
That simple rule governs much of the infectious behavior of viruses of various genome types, as we shall see. The seven genomic classes of viruses are:
* The word "only" is ironic here. All life forms on Earth use the same genetic system — double-stranded DNA — to store and use genetic information. While viruses aren't alive, they employ a much broader range of genetic storage.
| Type | Explanation | |
|---|---|---|
| I | dsDNA | double-strand ±DNA |
| II | ssDNA → dsDNA | single-strand (+)DNA with a double-strand DNA intermediate |
| III | dsRNA | double-strand ±RNA |
| IV | (+)ssRNA → (-)ssRNA | single-stranded (+)RNA with a single-strand (-)RNA intermediate (see VI) |
| V | (-)ssRNA | single-strand (-)RNA |
| VI | ssRNA-RT → DNA/RNA | single-strand (+)RNA with single-strand (-)DNA intermediate (see IV) |
| VII | ds, gapped DNA | ds DNA with gaps to be repaired in the nucleus of the host cell |
Symbols used on this page
In the sections below, we'll use this symbol

to represent a viral protein translated from an mRNA by the host ribosome, and this symbol

to represent a newly-constituted virus particle with its genetic material inside, surrounded by a protein or protein-membrane coat.
We'll use green squiggles to represent RNA, light green for a (+) strand of RNA. The (+) strand is just the strand oriented in the direction of the code for the protein to be translated. The (-) strand is its complement:
![]()
The (-) RNA strand will be a darker green squiggle:
![]()
DNA will be rendered in blue. A (+) DNA strand will be a light blue squiggle like this:
![]()
And a (-) DNA strand will be a darker blue squiggle:
![]()
These coloring schemes are roughly in keeping with those used in a couple of major textbooks on virology and immunology, and they'll be used throughout the pages on this site on viruses.
About polymerases
There are four basic kinds of nucleic-acid polymerases, enzymes that read the sequence of one strand of a nucleic acid polymer (RNA or DNA) and produce the base-paired complement to that strand.
- DNA-dependent DNA polymerase (DNAp) makes a complement strand of DNA from a DNA template.
- DNA-dependent RNA polymerase (RNAp) makes an RNA from a DNA strand. In most cases, we're talking about making an mRNA from the (-) or antisense strand of a DNA duplex in a cell.
- RNA-dependent DNA polymerase (RdDp) makes a DNA strand from its complement RNA strand.
- RNA-dependent RNA polymerase (RdRp) makes a complement RNA strand from another RNA strand.
The word "directed" is often substituted for "dependent" in some writing.
Overview: Viral genome types
Here is a graphical overview of the seven Baltimore virus classes. Notice that, one way or another, all classes must either contain or transcribe a messenger RNA that encodes its proteins. If mRNAs cannot be made, the virus can't be reproduced.

Type I: double-strand DNA
We know that in all living organisms (we don't consider viruses to be alive), DNA carries the instructions for proteins and a few RNAs, all that is needed to support cellular life. The (-) or non-coding strand of DNA is used to form a complement (+) RNA strand, the "message." That messenger RNA (mRNA) will be translated into the protein it encodes on the ribosomes in the cytoplasm of the host cell.
$$\text{-DNA } \: \rightarrow \: \text{ mRNA } \: \rightarrow \: \text{ protein}$$
The genetic material of double-stranded DNA viruses is a mimic of the dsDNA already present in the nucleus of any cell. If it can get into the nucleus, it can be transcribed by the host RNA polymerase (RNAp).
Generally, access to the nucleus is through a channel called the nuclear pore complex (NPC), and each virus has its own unique way of getting through this selective gateway. Some bypass it and get through the nuclear membrane another way. A nice review of these pathways can be found in: Fay & Pante, Frontiers in Microbiology (6), 467 (2015).
Once in the nucleus, viral dsDNA is transcribed and duplicated by the host DNA polymerase (DNAp), generating fresh genetic and protein building blocks for new viruses. A schematic of the process is shown below.
Some viruses come with their own RNAp and can make transcripts outside of the nucleus. Others can coax host nuclear RNAp out into the cytoplasm to trascribe their DNA to mRNA. The virome is highly diverse. dsDNA genomes can be circular (like a plasmid), linear and linear with covalently-linked ends (terminated loops).
Some dsDNA viruses encode their own DNA polymerase, and therefore don't
Examples of dsDNA viruses are:
- adeno viruses (adenoviridae);
- herpes viruses (herpesviridae);
- papilloma viruses (papillomaviridae), which can cause cancers like cervical cancer;
- polyoma viruses (polyomaviridae)
- pox viruses (poxviridae), which cause diseases like chicken pox and smallpox.
dsDNA viral replication schematic

Type II: Single-strand (+) DNA
In this genome class, the viral code is contained on a single (+) strand of DNA. The genomes of ssDNA viruses are rather spare, usually encoding a single coat protein (CP) and a replication associated protein (Rep), an endonuclease*. Most ssDNA viruses have circular genomes which are transcribed by host RNAp (with various kinds of help from the Rep enzyme) in a process called rolling circle replication, in which new transcripts come off the circular template as the polymerase works around. The resulting linked transcripts are later processed for translation. Because host RNAp is involved, the viral genetic material must access the host cell nucleus.
ssDNA viruses are ubiquitous on Earth and infect all kingdoms of life: prokaryotes, archea and eukaryotes. In humans, parvovirus is a common ssDNA pathogen that can have bad consequences for people with certain underlying conditions such as sickle-cell disease or pregnancy, but for the most part don't have any lasting effects. When parvoviruses replicate and package their genomes, they can package (-) or (+) strand DNA, and each is fully capapble of infecting and replicating in a new host cell.
Nucleases are enzymes that cleave nucleic acid chains (RNA, DNA). Exonucleases break the bonds of the end-terminal nucleotides, while endonucleases cleave in the interior of the chain.
ssDNA viral replication schematic
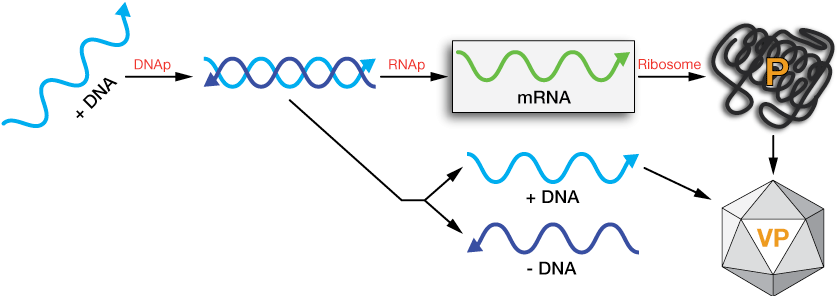
Type III: Double-strand RNA
Double-strand RNA viruses contain base-paired RNA. When pulled apart, the (+) strand(s) can be used directly as mRNA to make viral proteins and form new particles. The (+) strand can also be converted, using RNA-dependent RNA polymerase (RdRp) into more dsRNA.
Host cells do not contain RdRp, so that, along with its genetic code, must be carried inside of the virus particle. Eukaryotic host cells contain one or more defenses against ds RNA, even going so far as to trigger cell death (apoptosis). To circumvent these defenses, the formation of mRNAs from the viral dsRNA is often done right inside the intact particle (inside the capsid) while it is in the host cell. Sometimes the RdRp enzymes even form a part of a pore through the viral capsid, churning out mRNAs from the protected dsRNA inside.
dsRNA viral replication schematic
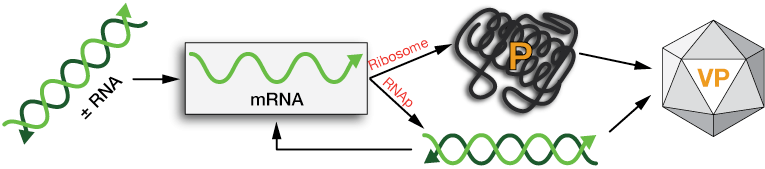
Type IV: Single-strand (+) RNA
with -RNA intermediate
Single-strand (+) RNAs are messenger RNAs, and thus ready for translation to proteins once released into the cytoplasm of the host cell. An RNA polymerase (RNAp) contained in the capsid copies the (+) strands to (-) strands, which are in turn copied to make more (+) strands.
A wide variety of pathogenic viruses are (+) RNA viruses. They include
- Corona viruses, which cause SARS, COVID-19 and MERS;
- Picorna viruses, some of which cause polio;
- Atroviruses & Caliciviruses, which cause gastroenteritis;
- Flaviviruses, which cause Yellow fever, West Nile fever, Dengue fever, Hepatitis C and other illnesses;
- Togaviruses, which cause rubella.

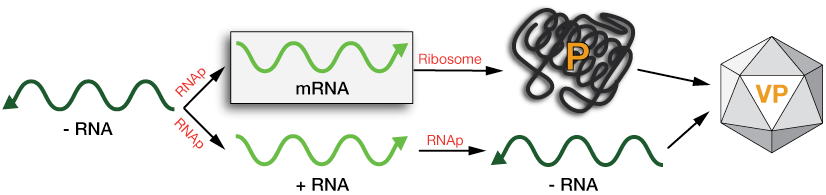
Type V: Single-strand (-) RNA
The single-strand (-) RNA's, which can come in a single piece or several fragments, depending on the virus, must be copied to a (+) strand in order to be translated into protein or into more (-) strands to build more viruses.
Host cells do not possess the polymerase, an RNA-dependent RNA polymerase (RNAp), that does this copying; it has to be supplied, as an intact enzyme, inside the viral capsid. When new viruses are formed, a new copy of the RNAp is packaged inside the VP with the new (-)RNAs.
The RNA-dependent RNAp can produce either translation-ready mRNAs for protein construction, or non-messenger (+) RNAs that are used to make new (-) RNAs for new viruses.
Flu & reassortment
The influenza viruses are good examples of (-) RNA viruses that have segmented genomes. Flu virus particles have a genome that consists of eight segments of 10,000 - 15,000 bases (10-15 Kb) each. One of the reasons that flu virus has a higher rate of phenotypic variablity and a higher rate of formation of infectious variants than most other viruses is that its genome is susceptible to reassortment. It works like this (see figure): Imagine that two different flu viruses infect the same cell. In the figure, one has red RNA segments and the other blue.
Because those segments can mix inside of a cell, when they are repackaged into VPs, reassortment or mixing of the segments can occur, creating functional viruses with properties that may be different.

ss -RNA viral replication schematic
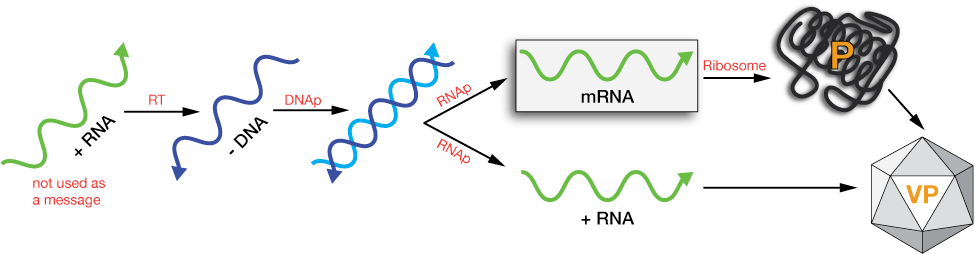
Type VI: Single (+) strand RNA with (-) DNA intermediate
This is the genome of the retroviruses, like HIV, which can insert a copy of their genome into the host DNA. A single-strand of RNA is copied by reverse transcriptase (RT), an enzyme carried by the virus that does not exist in cells. These viruses are called retroviruses because the can perform the reverse operation of transcription, copying an RNA strand to a DNA strand.
The DNA copy is duplicated by the cellular machinery in the nucleus to form a double helix, which is then inserted into the host-cell genome.
After insertion, the viral DNA can be quiescent (quiet) for a time, then transcribed into more viral RNA to form new viral proteins and RNAs to be packaged into new virions. Retroviruses must contain both RT and an enzyme called an integrase, which inserts the viral DS-DNA into the host DNA. Retroviruses are thought to have infected vertebrates for 450 million years or so.
retrovirus replication schematic
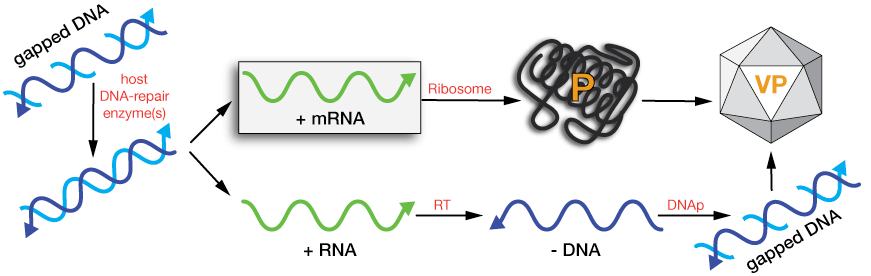
Type VII: Gapped DNA
This genome class compares most closely with that of dsDNA viruses (type I). In order for it to produce mRNAs in the host cell, the gaps must be repaired to yield intact dsDNA that can then be transcribed.
This involves a series of steps, some of which are unique to gapped-DNA viruses and some of which strongly resemble the replication of retoroviral (type xx) viruses: The protein and RNA must be removed, the missing part(s) of the (+) strand of DNA must be repaired to produce a dsDNA that can be transcribed. Then, in order to build more viruses, both new capping proteins and RNAs must be made and attached, and some process to build the gapped (+) strand from an intact (-) strand must be deployed.
Schematically, gapped-DNA genomes look something like the figures below. In the upper (linear-DNA) figure, the (-) strand is intact and has a small protein attached to its 5' end. An RNA is attached to the 5' end of the partial (+) DNA. In hepatitis B virus, the gapped-dsDNA genome is circular – like an incomplete plasmid.

Gapped DNA viral replication schematic
Virome
The virome revers to the collection of all viral genomes, or even just viruses, that live within an organism, a set of organisms or a region on Earth. It is used rather loosely in context.
We can refer, for example, to the global virome (all viruses on Earth) or the human virome, the viruses that infect humans.

![]()
xaktly.com by Dr. Jeff Cruzan is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. © 2012, Jeff Cruzan. All text and images on this website not specifically attributed to another source were created by me and I reserve all rights as to their use. Any opinions expressed on this website are entirely mine, and do not necessarily reflect the views of any of my employers. Please feel free to send any questions or comments to jeff.cruzan@verizon.net.